





















Universität Bielefeld › Technische Fakultät › NI
Search
Unsupervised Kernel Regression (UKR)
In general, the domain of manifold learning addresses the problem of finding a lower dimensional (latent) representation 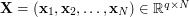 of a set of observed data
of a set of observed data 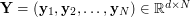 and the corresponding functional relationship
and the corresponding functional relationship 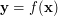 .
.
Unsupervised Kernel Regression (UKR) is a recent approach to learning non-linear continuous manifold representations. It has been introduced as an unsupervised formulation of the Nadaraya-Watson kernel regression estimator by Meinecke, Klanke et al. and further developed by Klanke. It uses the Nadaraya-Watson estimator as mapping 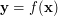 between latent and observed space:
between latent and observed space:
![\[ \mathbf{y} = f(\mathbf{x}) = \sum_{i=1}^N{\mathbf{y}_i \frac{\mathbf{K}_{\mathbf{H}}(\mathbf{x} - \mathbf{x}_i)} {\sum_j \mathbf{K}_{\mathbf{H}}(\mathbf{x} - \mathbf{x}_j)}} \]](/files/tex/5139d4139fd7b832423e706623145f2e2a1a4026.png) |
(1) |
This estimator realises a smooth, continuous generalisation of the functional relationship between two random variables  and
and 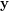 described by given data samples
described by given data samples 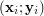 . Here,
. Here, 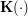 is a density kernel like Gaussian or Quartic and
is a density kernel like Gaussian or Quartic and 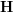 contains the corresponding bandwidths. Whereas the choice of the kernel is of relatively low importance, the bandwidth plays a more crucial role in the original estimator.
contains the corresponding bandwidths. Whereas the choice of the kernel is of relatively low importance, the bandwidth plays a more crucial role in the original estimator.
UKR now treats (1) as a mapping from the latent space of the represented manifold to the original data space in which the manifold is embedded and from which the observed data samples 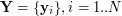 are taken. The associated set
are taken. The associated set 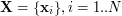 now plays the role of the input data to the regression function (1). Here, they are treated as \textit{latent parameters} corresponding to
now plays the role of the input data to the regression function (1). Here, they are treated as \textit{latent parameters} corresponding to 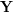 . As the scaling and positioning of the
. As the scaling and positioning of the 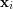 's are free, the formerly crucial bandwidth parameter
's are free, the formerly crucial bandwidth parameter 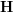 becomes irrelevant and we can use unit bandwidths.
becomes irrelevant and we can use unit bandwidths.
The UKR regression function 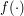 thus can be denoted as
thus can be denoted as
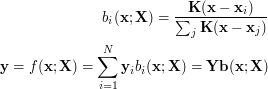 |
(2) |
where 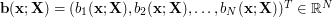 is a vector of basis functions representing the effects of the kernels parametrised by the latent parameters.
is a vector of basis functions representing the effects of the kernels parametrised by the latent parameters.
The training of the UKR manifold, or the adaptation of 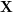 respectively, then can be realised by gradient-based minimisation of the reconstruction error
respectively, then can be realised by gradient-based minimisation of the reconstruction error
![\[ R(\mathbf{X}) = \frac{1}{N}\sum_{i} \parallel \mathbf{y}_i - f(\mathbf{x}_i; \mathbf{X}) \parallel^2 = \frac{1}{N}\parallel\mathbf{Y} - \mathbf{YB}(\mathbf{X})\parallel^{2}_{F} \]](/files/tex/d5b04a8d2bd9563ffcfdbb31fea4198ef378e0cb.png) |
(3) |
where 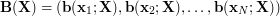 .
.
To avoid the trivial minimisation solution 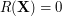 by moving the
by moving the 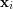 infinitely apart, several regularisation methods exist. Here, the most outstanding to mention is UKR's ability of very efficiently performing a leave-one-out cross-validation, that is, reconstructing each
infinitely apart, several regularisation methods exist. Here, the most outstanding to mention is UKR's ability of very efficiently performing a leave-one-out cross-validation, that is, reconstructing each 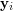 without using itself. To this end, in the calculation of
without using itself. To this end, in the calculation of 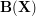 (cf. (3)), the only additional step is to zero its diagonal before normalising the column sums to 1.
(cf. (3)), the only additional step is to zero its diagonal before normalising the column sums to 1.
|
|
|||||||||
 |
 |
 |
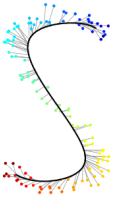 |
 |
|||||
| n=0 | n=5 | n=10 | n=20 | n=100 | |||||
|
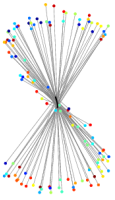
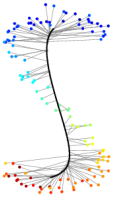
Fig.1: Progress of UKR training of a "noisy S" after n=0,5,10,20,100 gradient steps. Depicted are the reproduction of the one-dimensional UKR latent space (black line) and the corrsponding training data points (points, colours visualise the value of the 1D-latent parameter). |
|||||||||
Note that, for a preselected density kernel, the reconstruction error (3) only depends on the set of latent parameters 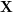 . Thus, minimising
. Thus, minimising 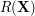 w.r.t.
w.r.t. 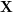 optimises both the positioning of the latent parameters
optimises both the positioning of the latent parameters 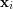 's and the shape of the manifold at the same time. In this feature, UKR differs from many other manifold representations which usually require alternating projection and adaptation steps.
's and the shape of the manifold at the same time. In this feature, UKR differs from many other manifold representations which usually require alternating projection and adaptation steps.
As gradient descent often suffers from getting stuck in poor local minima, an appropriate initialisation is important. Here -- depending on the problem -- i.e. PCA , Isomap or Local Linear Embedding (LLE) are usually good choices. These methods themselves are quite powerful in uncovering low-dimensional structures in data sets. In contrast to UKR indeed, PCA is restricted to linear structures and Isomap as well as LLE do not provide smooth mappings into the data space.
An inverse mapping 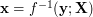 from data space to latent space is not directly supported in UKR. Instead, one may use an orthogonal projection to define a mapping
from data space to latent space is not directly supported in UKR. Instead, one may use an orthogonal projection to define a mapping 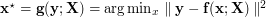 which approximates
which approximates 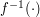 .
.
UKR is presented in more detail in:
- Login to post comments
